Single-cell sequencing represents a paradigm shift in genomics, offering unprecedented insights into the complexities of cellular behavior and function. This rapidly advancing field hinges on the ability to analyze individual cells, uncovering heterogeneities and rare cell populations that elude bulk sequencing methods. The accuracy of single-cell sequencing critically depends on the quality of sample preparation, emphasizing the importance of precise cell counting and sample assessment before sequencing.
The quality of single nuclei is paramount in single-cell genomics, as it directly influences the reliability and accuracy of sequencing data. High-quality nuclei samples ensure that the genetic information derived is representative and free from artifacts that could skew results. This section would delve into the challenges and techniques involved in preparing high-quality nuclei samples, highlighting why meticulous preparation is crucial for successful single-cell sequencing.
Advances in automated cell counting technology, such as the LUNA-FX7™ Automated Cell Counter, have streamlined the process of nuclei quality assessment. These sophisticated devices utilize fluorescent dyes and imaging tools to provide an accurate, reliable measure of nuclei quality.
Automated Cell Counter such as LUNA-FX7™ is an advanced platforms with robust capabilities for comprehensive analysis. These systems seamlessly capture both brightfield and fluorescent images that are then analyzed to obtain valuable data such as size, viability, and cell count using fluorescent signals.
To evaluate the quality of isolated nuclei, one can utilize the average cell size data provided by these systems. For instance, nuclei from which the membrane has been removed are measured to be significantly smaller in size compared to intact cells. This is because the removal of the cell membrane and cytoplasm leads to a reduction in cell size by several micrometers. In particular, the LUNA-FX7™ extract cell size information from the brightfield image. As a result, there’s minimal distortion of cell size due to fluorescence intensity that might accompany calculations from fluorescent images. This ensures a more reliable cell size measurement when assessing the quality of isolated nuclei.
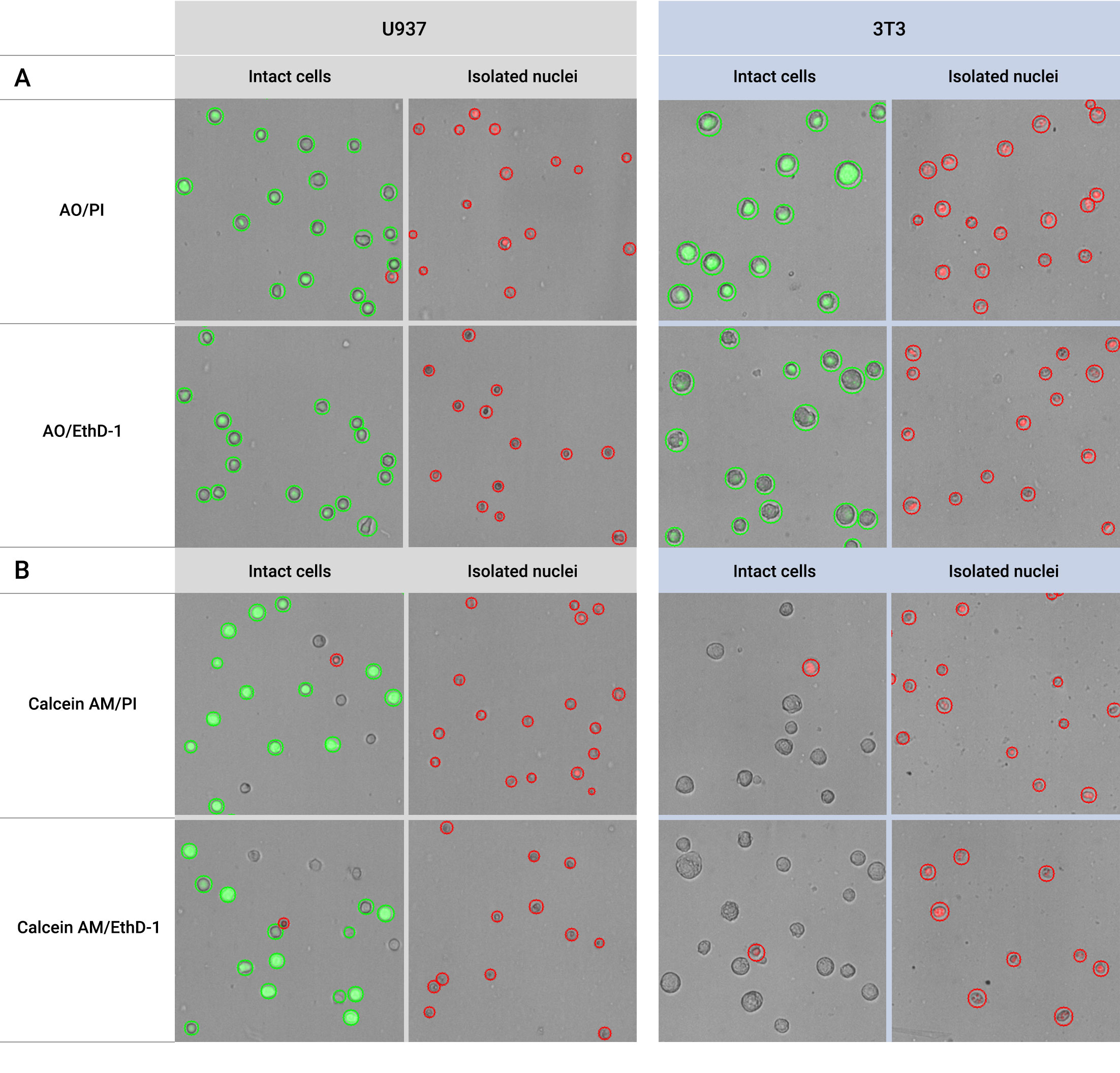
Another useful data for nuclei quality assessment is cell viability using the fluorescent dyes. As indicated in results in Figure 1, when combined with AO/PI or AO/EthD-1, healthy intact cells exhibit cell viability close to 100%. In contrast, well-separated nuclei display cell viability close to 0%. Notably, automated cell counters like LUNA-FX7™ enable accurate quantification of isolated nuclei cell concentration when stained with these dyes.
In conclusion, the integration of automated cell counting technology in single-cell genomics has been a game-changer, enhancing the accuracy and reliability of research outcomes. The LUNA-FX7™ Automated Cell Counter exemplifies this advancement, offering researchers a powerful tool for assessing the quality of nuclei samples. As the field of genomics continues to evolve, such technological innovations will remain pivotal in driving forward our understanding of the cellular world.
Fluorescence Dyes and Automated Cell Counters: Towards Reliable Nuclei Quality in Single Cell Genomics. https://logosbio.com/application_notes/fluorescent-dyes-and-automated-cell-counters-towards-reliable-nuclei-quality-in-single-cell-genomics//
For more information about the LUNA-FX7™ Automated Cell Counter, visit https://logosbio.com/luna-fx7/.