Dr. Dennis Nurjadi discusses the cell counting technology that has brought his team accuracy and reproducibility
In this article, we hear from Dr. Dennis Nurjadi, Head of Molecular Bacteriology at the University Hospital Heidelberg, about his team’s work on Staphylococcus aureus and the importance of understanding its mechanisms in order to develop novel strategies to decolonize high-risk patients and prevent S. aureus infections. Here, Nurjadi explains the current challenges labs face when quantifying bacteria and how Logos Biosystems’ QUANTOM Tx addresses these issues by producing reliable, accurate, and reproducible quantifications.

The QUANTOM Tx at the University Hospital Heidelberg © Dr. Dennis Nurjadi
What are the overall aims of your microbial research?
Our group works on Staphylococcus aureus, focusing on the pathogen host-interaction in terms of colonization and infection, but also on the molecular mechanisms of antibiotic resistance.
We are currently working on the interaction between Staphylococcus aureus and keratinocytes and are looking at the immunological response, how skin cells/ keratinocytes sense and differentiate between pathogen and commensal organisms. For this, we use an in vitro infection assay to study the reaction of keratinocytes to various microbial stimuli (e.g. viable and dead bacteria), in terms of immune signaling.
What are the intended impacts and clinical implications?
We would like to understand the mechanism behind S. aureus colonization better and why commensal organisms on the skin are tolerated by the skin’s immune defense mechanisms. Besides, individuals with S. aureus colonization are at risk of developing S. aureus infections after invasive procedures such as surgery, dialysis. A better understanding of the host-pathogen interplay may contribute to develop novel strategies for the decolonization of high-risk patients and prevent S. aureus infections.
Do you have any preliminary results?
The results are not published yet, but we can share some data. In our experiments, we saw differences in immune response, depending on the infection with inactivated bacteria or viable bacteria. In some of the assays, we saw some heterogeneity in the internalization of the bacteria, which may be a result of a heterogeneous bacterial population, which are on various growth stages or even a mixture of dead and viable bacteria.
What are the current challenges you encounter in your research?
Quantification of bacteria usually relies on turbidity measurements, meaning no differentiation between dead and viable cells. To obtain reproducible and reliable results, the quantification is one of the most crucial steps, as misquantification can lead to divergent and non-reproducible results, resulting in variations and deviations.
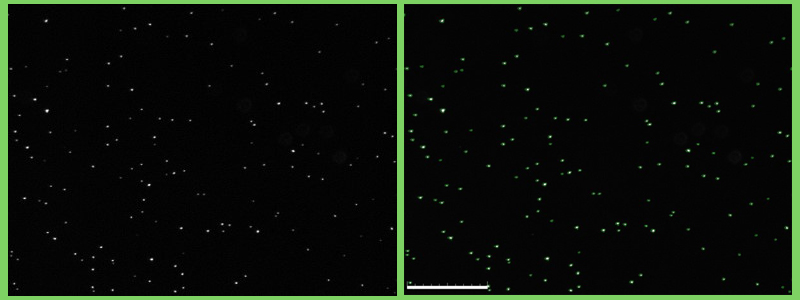
Fluorescent cells (Staphylococcus epidemidis) visualized using the QUANTOM Tx (L: Untagged, R: Tagged) © Dr. Dennis Nurjadi
How do you use the QUANTOM Tx Microbial Cell Counter?
We used the QUANTOM Tx Microbial Cell Counter first to quantify the total amount of bacteria and then to quantify viable bacteria with the viable staining. Then the bacterial suspension is diluted to the desired concentration. We mainly use this equipment to quantify bacteria for infection assays.
Its key benefit has been in providing correct quantification of bacteria, thus minimizing variations in the bacterial suspension. With this, we were able to calculate the number of viable bacteria in relation to total bacterial cells. We were also able to quantify and visualize the bacterial cells. This would allow us to verify the cell morphology and to identify possible contaminants.
What do you see for your research in the future?
Regarding the QUANTOM Tx Microbial Cell Counter, we will use this equipment to study mechanisms of plasmid transfer (i.e., calculating frequency and efficiency), where reliable cell count is essential. Another possible use of this machine would be for a bacterial killing assay, for example, to investigate the antibacterial effect of substances.
The time factor is still a challenge; the preparation time is significantly more than the turbidity-based estimation of CFU in a bacterial suspension. This machine still has a lot of potential which has not been fully explored in our lab. As far as future developments go, it would be great if it were possible to incorporate two different stains (fluorescent channels) to study interaction or to quantify multiple species in one experiment.










